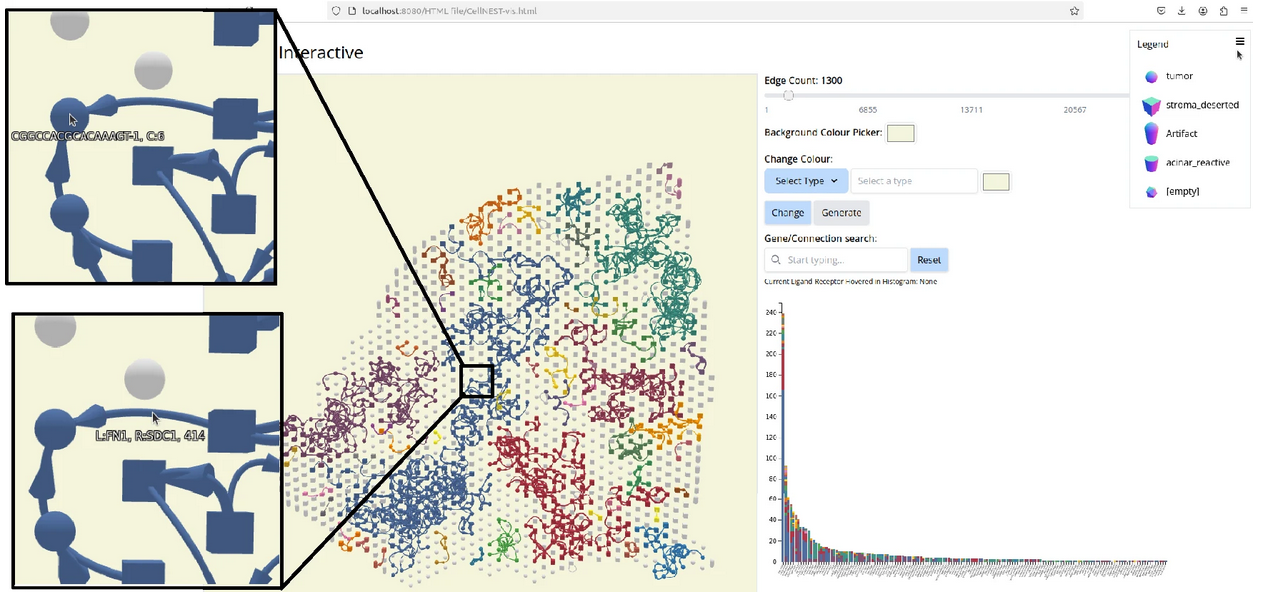
Nature Methods, 2025
Zohora FT*, Paliwal D*, Flores-Figueroa E, Li J, Gao T, Notta F & Schwartz GW
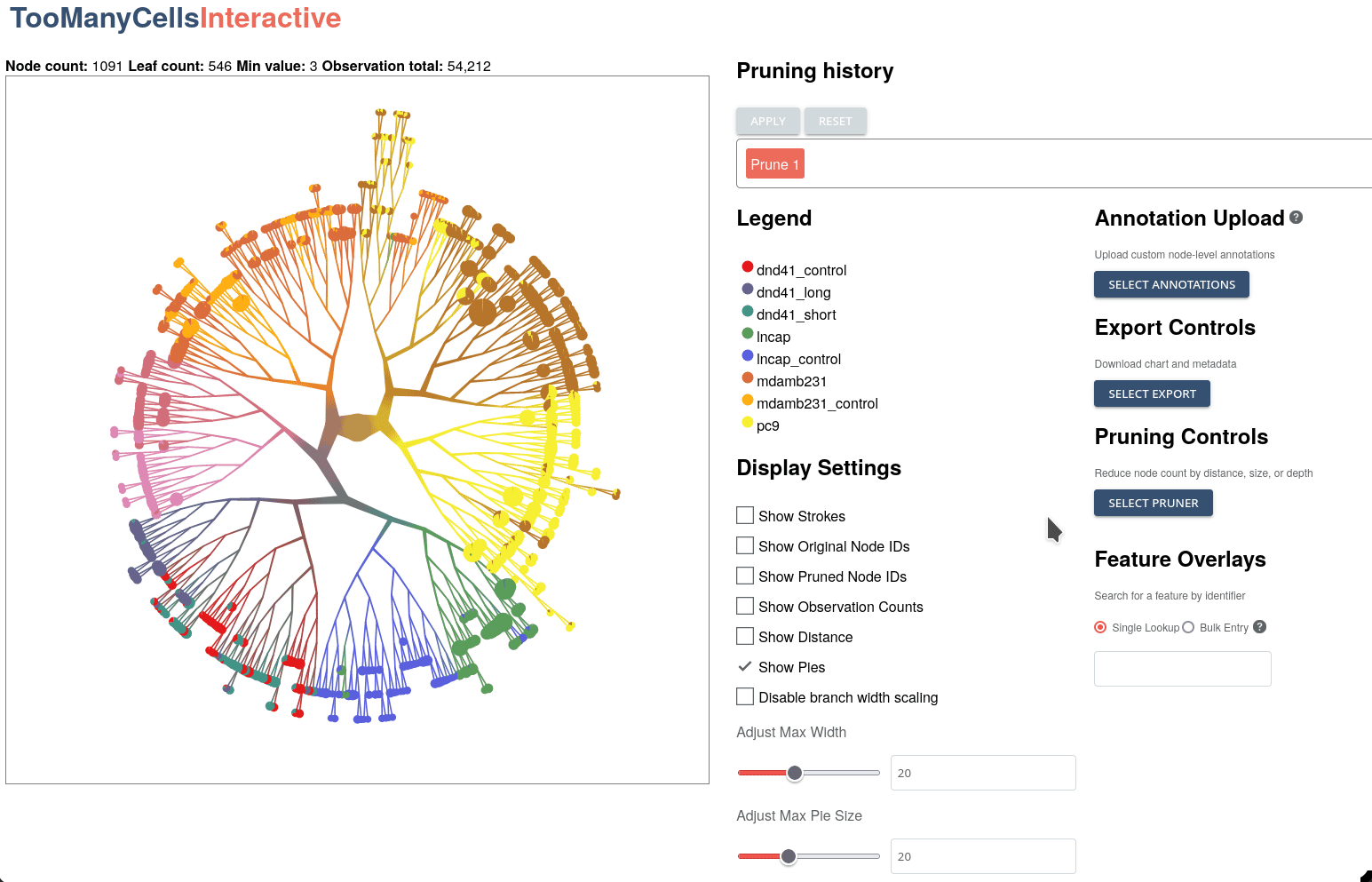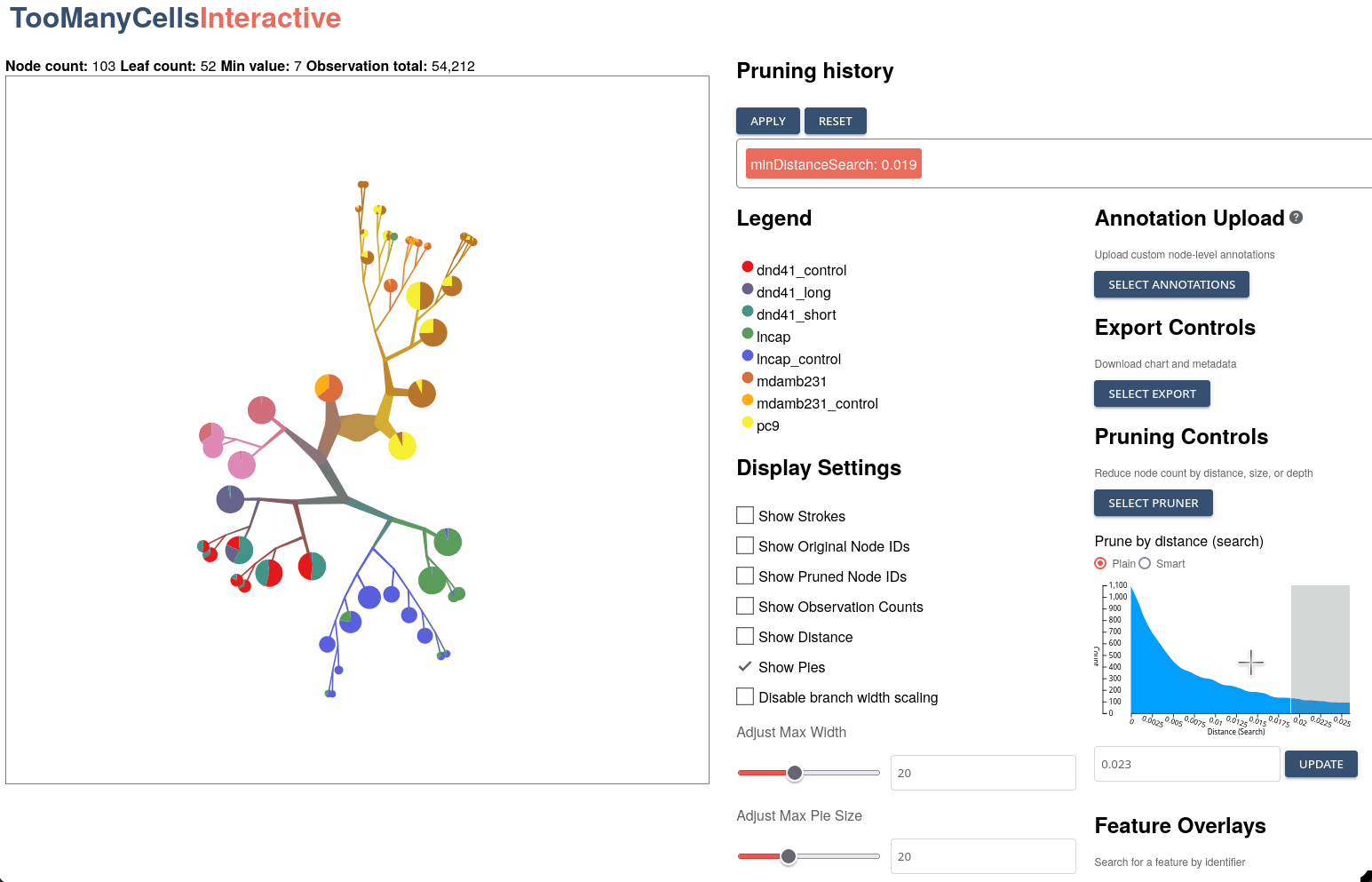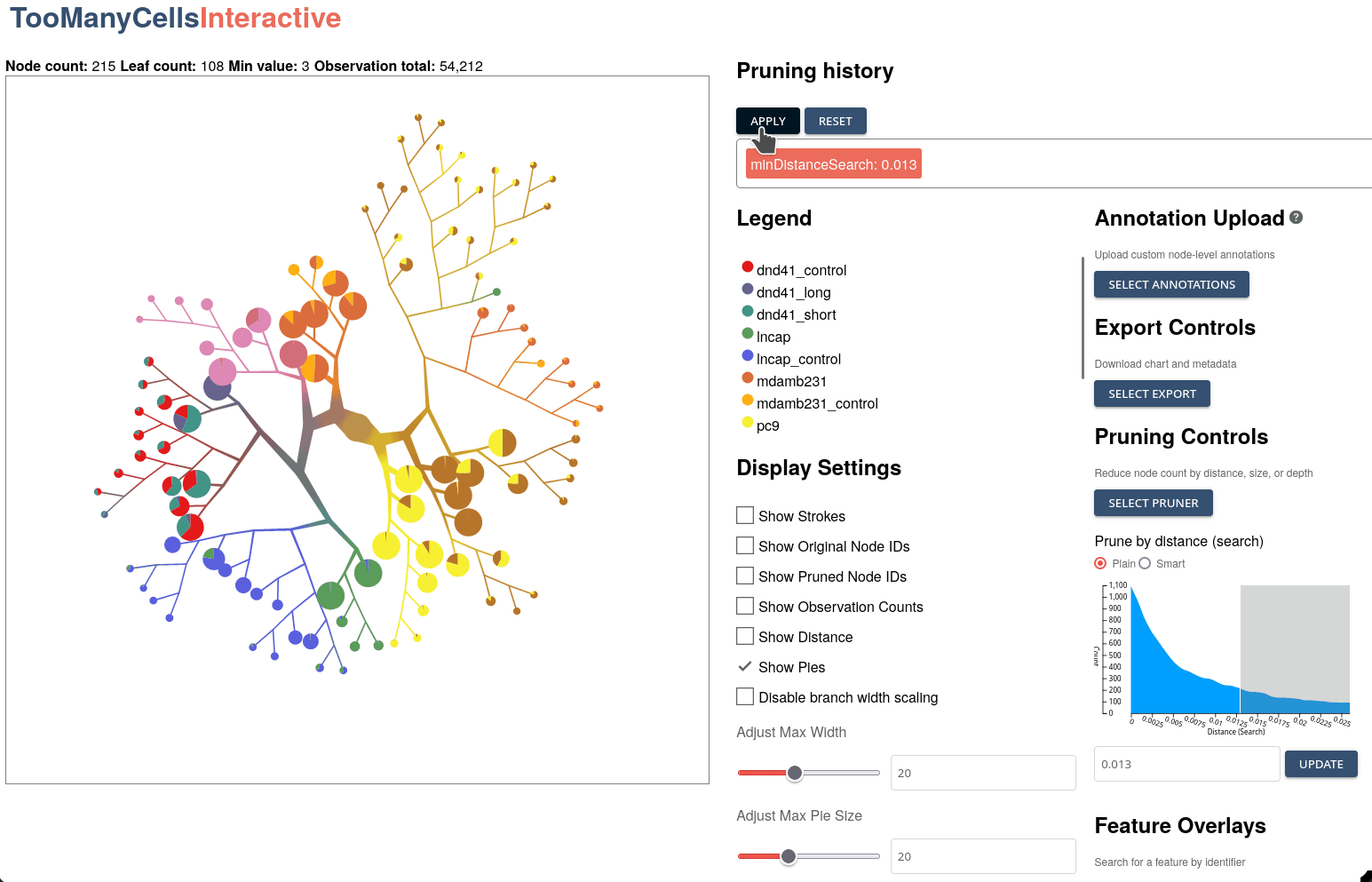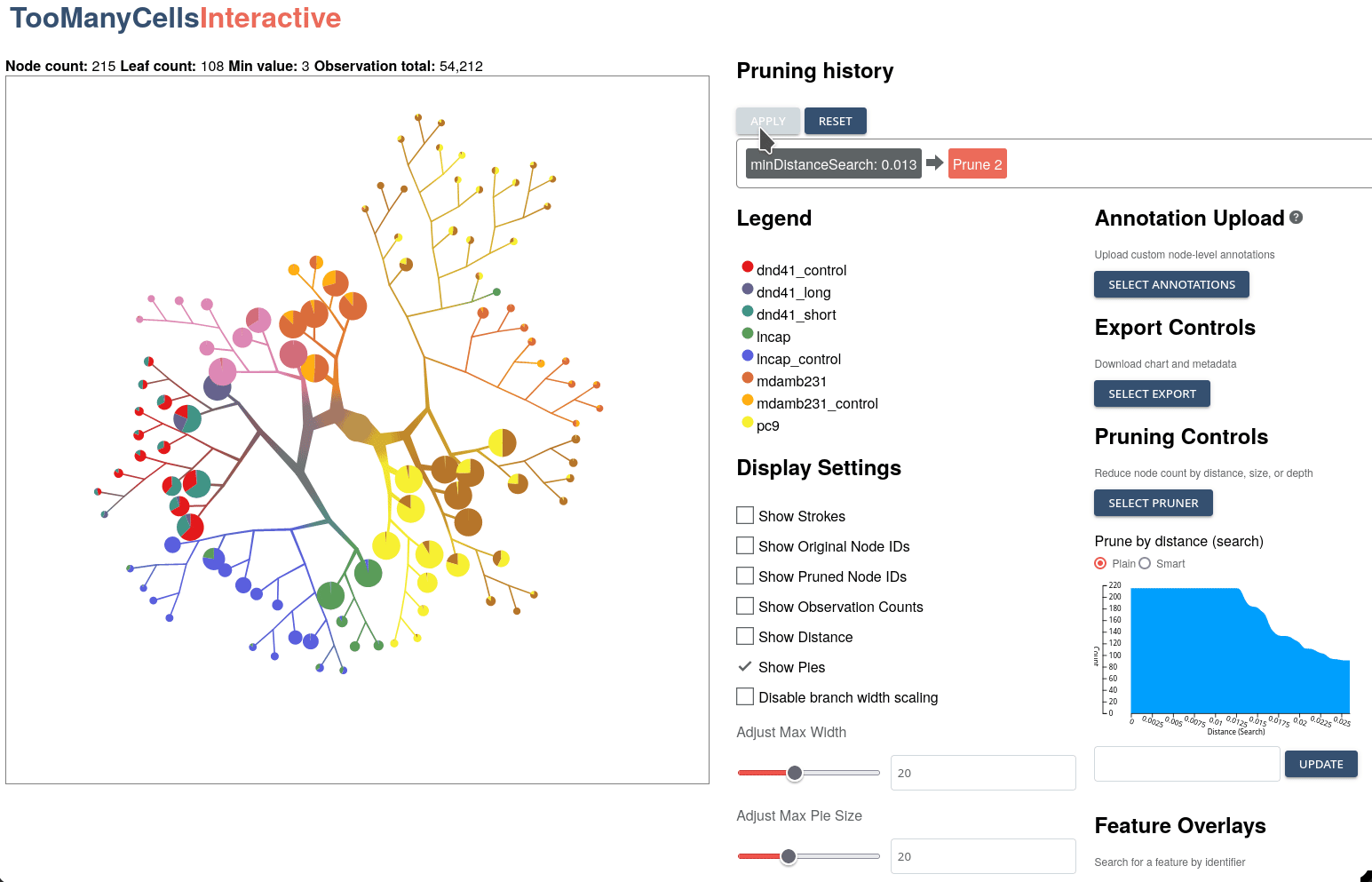
GigaScience, 2024
Klamann C*, Lau, CJ*, Ruiz-Ramírez J, and Schwartz GW
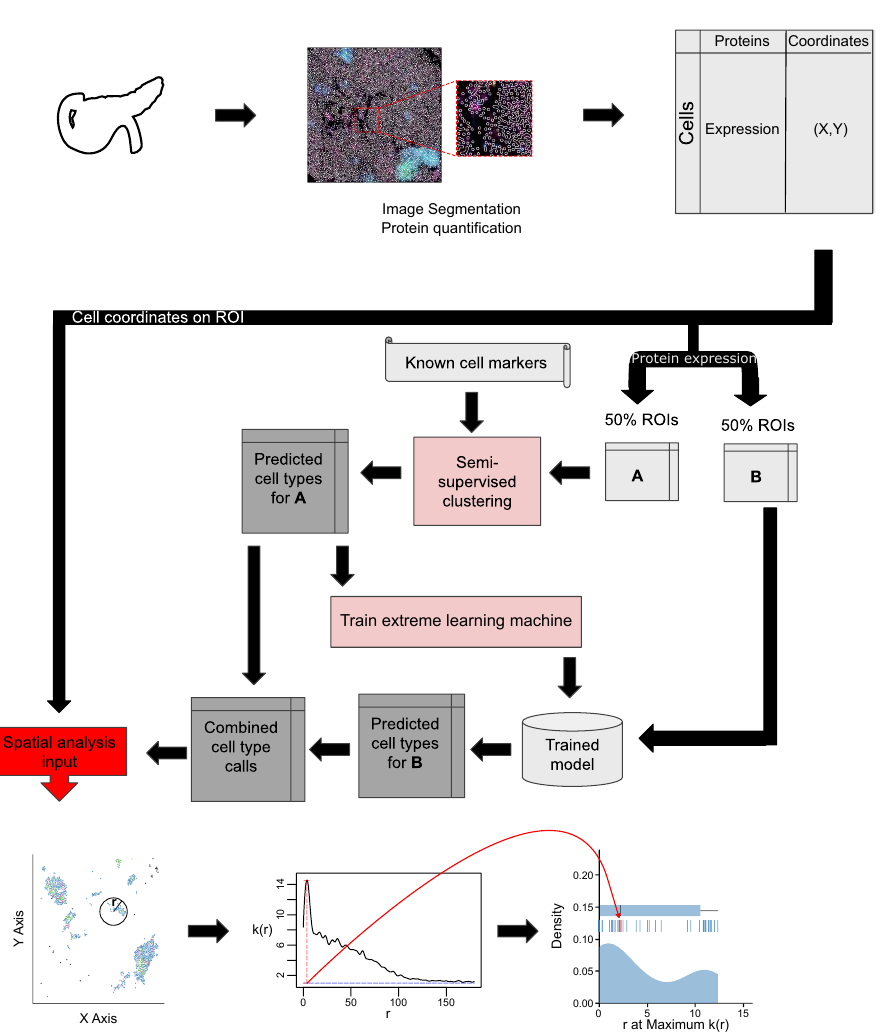
Nature Communications, 2024
Mongia A, Zohora FT, Burget NG, Zhou Y, Saunders DC, Wang YJ, Brissova M, Powers AC, Kaestner KH, Vahedi G, Naji A, Schwartz GW^, and Faryabi RB^.
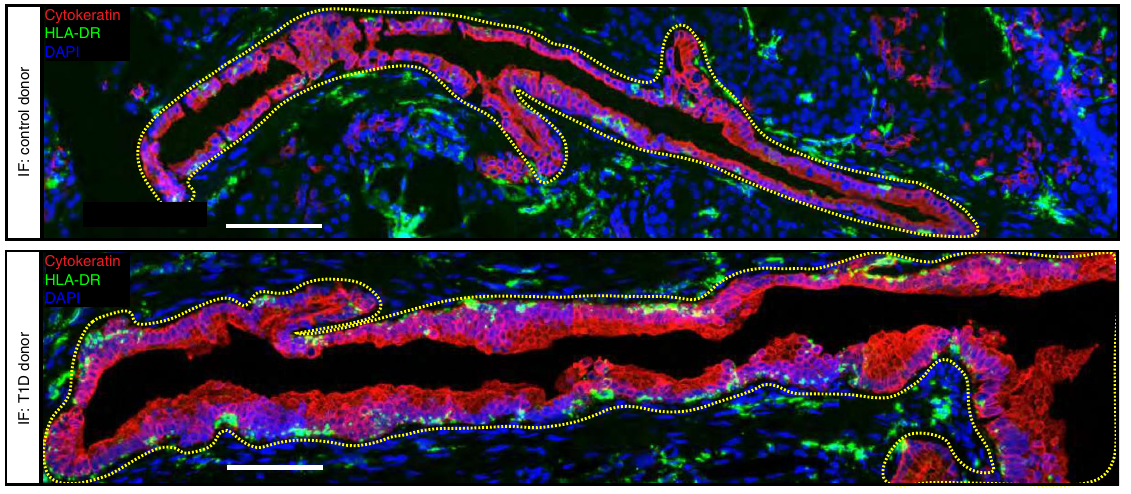
Nat Metab, 2022
Fasolino M*, Schwartz GW*, Patil AR, Mongia A, Golson ML, Wang YJ, Morgan A, Liu C, Schug J, Liu J, Wu M, Traum D, Kondo A, May CL, Goldman N, Wang W, Feldman M, Moore JH, Japp AS, Betts MR; HPAP Consortium, Faryabi RB, Naji A, Kaestner KH, Vahedi G.
bioRxiv 2025
Dong ZF, McIntosh C & Schwartz GW
Nature Methods 2025
Zohora FT*, Paliwal D*, Flores-Figueroa E, Li J, Gao T, Notta F & Schwartz GW
Science Translational Medicine 2025
Gower M, Li X, Aguilar-Navarro AG, Lin B, Fernandez M, Edun G, Nader M, Rondeau V, Arruda A, Tierens A, Eames Seffernick A, Pölönen P, Durocher J, Wagenblast E, Yang L, Lee HS, Mullighan CG, Teachey D, Rashkovan M, Tremblay CS, Herranz D, Itkin T, Loghavi S, Dick JE, Schwartz G, Perusini MA, Sibai H, Hitzler J, Gruber TA, Minden M, Jones CL, Dolgalev I, Jahangiri S & Tikhonova AN
Nature Communications 2024
Perlman BS, Burget N, Zhou Y, Schwartz GW, Petrovic J, Modrusan Z & Faryabi RB
GigaScience 2024
Klamann C*, Lau, CJ*, Ruiz-Ramírez J, and Schwartz GW
bioRxiv 2024
Zohora FT, Flores-Figueroa E, Li J, Paliwal D, Notta F, and Schwartz GW
Nature Communications 2024
Mongia A, Zohora FT, Burget NG, Zhou Y, Saunders DC, Wang YJ, Brissova M, Powers AC, Kaestner KH, Vahedi G, Naji A, Schwartz GW^, and Faryabi RB^.
bioRxiv 2023
Klamann C, Lau C, Schwartz GW
bioRxiv 2023
Mongia A, Saunders DC, Wang YJ, Brissova M, Powers AC, Kaestner KH, Vahedi G, Naji A, Schwartz GW^, Faryabi RB^.
Nat Metab 2022
Fasolino M*, Schwartz GW*, Patil AR, Mongia A, Golson ML, Wang YJ, Morgan A, Liu C, Schug J, Liu J, Wu M, Traum D, Kondo A, May CL, Goldman N, Wang W, Feldman M, Moore JH, Japp AS, Betts MR; HPAP Consortium, Faryabi RB, Naji A, Kaestner KH, Vahedi G.